Integrons have been recognised as important contributors to the acquisition and dissemination of antibiotic resistance in Gram-negative bacteria. In a collection of 19 multi-antibiotic resistant Gram-negative clinical isolates, 47 per cent (9/19) of strains were found to contain one or more integron, using a polymerase chain reaction (PCR) based screening method. Resistance gene cassettes within the integrons were amplified, sequenced and characterised. Antibiotic susceptibility testing demonstrated that resistance phenotypes correlated with the resistance conferred by gene cassettes identified. PCR-screening for integrons and gene cassettes provides a rapid technique for the identification of genetic determinants of resistance in Gram-negative bacteria. Such screening could assist in guiding treatment regimens and complement existing antibiotic resistance surveillance programs by providing information on molecular mechanisms of both resistance and resistance dissemination. Commun Dis Intell 2003;27 Suppl:S103-S110.
Top of page
Introduction
Antibiotic resistance is a serious clinical problem worldwide. Acquisition of resistance genes in Gram-negative bacteria is facilitated by mobile genetic elements called integrons, which are associated with resistance plasmids and transposons.1 Integrons encode an enzyme, termed integrase, which allows them to capture antibiotic resistance gene cassettes (Figure).2,3 Over 80 cassettes have been identified to date, conferring resistance to almost all classes of antibiotic. The length of gene cassettes varies considerably from 262 base pairs (bp) to 1,549 bp,4,5 however, a common feature of all gene cassettes is a specific recombination site [termed 59-base element (59-be)], located downstream of the gene. The 59-be is recognised by the integron-encoded integrase (IntI),6 which enables the gene cassette to be inserted into the integron at a second recombination site (attI), located immediately upstream of the integrase gene (intI) (Figure).7,8,9,10 Gene cassette arrays in integrons can consist of up to nine cassettes,11 which are expressed from an upstream promoter (Figure).2,12,13 Integrons involved in antibiotic resistance can be divided into three classes, class 1, 2 and 3, based on the amino acid sequence of their respective integrases.
Figure. The insertion of a gene cassette into an integron
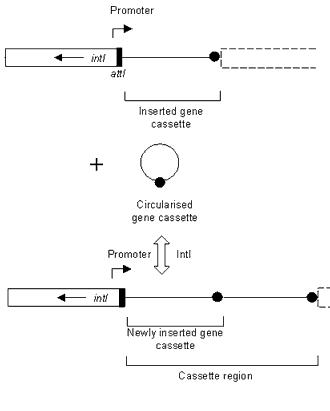
The preferential integration of gene cassettes at the attI recombination site (indicated by a black box), is catalysed by the product of intI, the integrase gene (open box). Two gene cassettes are indicated in the cassette region after the integration event. The filled circle represents the recombination site (59-be) of the gene cassette. A promoter for the expression of integrated gene cassettes, found upstream of the cassette region is also shown.
Integrons are prevalent amongst Gram-negative bacteria and have been associated with antibiotic resistance in clinical isolates.14,15,16,17,18,19 In a previous study we showed that integrons are significantly associated with multi-resistance in urinary isolates of Enterobacteriaceae.14 Investigations into the prevalence of integrons and characterisation of gene cassettes in clinical isolates provide information on the evolution of multiple-antibiotic resistant strains, the prevalence of antibiotic resistance genes and the molecular mechanisms of antibiotic resistance. This is important when considering strategies for effective antibiotic treatment of bacterial infections.
The present study investigates integrons and gene cassettes in a random selection of Gram-negative clinical isolates that were identified as multi-drug resistant. The presence of antibiotic resistance gene cassettes was correlated with the phenotypic antibiotic resistance profiles to evaluate the contribution of integrons to resistance.
Top of page
Materials and methods
Clinical isolates
Nineteen randomly selected multi-resistant strains of Gram-negative bacteria from a laboratory collection of clinical isolates were examined (Table 1). All isolates were obtained in 2000 and were considered multi-resistant if resistant to more than four classes of antibiotic. Escherichia coli Top 10 and E. coli NCTC 10418 were used as integron negative controls, while strains containing class 1, 2 and 3 integrons were also included as positive controls in all experiments.
Table 1. Integron status of bacterial strains studied
Strain |
Organism* |
Source of isolation† |
Integron |
| INSJ04 |
Proteus mirabilis |
N/A |
+ |
| INSJ07 |
Klebsiella sp. |
Urine |
– |
| INSJ08 |
Klebsiella sp. |
Urine |
– |
| INSJ09 |
Klebsiella sp. |
Urine |
– |
| INSJ10 |
Proteus mirabilis |
Urine |
+ |
| INSJ11 |
Klebsiella sp. |
Blood |
+ |
| INSJ12 |
Enterobacter cloaceae |
Wound |
– |
| INSJ14 |
Stenotrophomonas maltophilia |
Wound |
– |
| INSJ15 |
Acinetobacter baumanii |
Sputum |
– |
| INSJ16 |
Klebsiella sp. |
Urine |
+ |
| INSJ17 |
Acinetobacter baumanii |
Urine |
– |
| INSJ18 |
Acinetobacter baumanii |
Urine |
– |
| INSJ19 |
Acinetobacter baumanii |
Sputum |
– |
| INSJ20 |
Proteus mirabilis |
Urine |
+ |
| INSJ21 |
Escherichia coli |
Blood |
+ |
| INSJ22 |
Pseudomonas sp. |
Sputum |
– |
| INS95 |
Salmonella Typhimurium |
Stool |
+ |
| INSTR2 |
Citrobacter freundii |
Urine |
+ |
| INSTR5 |
Enterobacter cloaceae |
Urine |
+ |
Antibiotic susceptibility testing
Susceptibility to antimicrobial agents was determined using the Calibrated Dichotomous Sensitivity method.20 The antibiotics tested included: aminoglycosides (amikacin, gentamicin, kanamycin, netilmicin, streptomycin and tobramycin), ▀-lactams (ampicillin, augmentin, cefotaxime, cefotetan, cephalexin, imipenem and timentin), quinolones (nalidixic acid, norfloxacin), chloramphenicol, nitrofurantoin, sulphafurazole, tetracycline, and trimethoprim.
Detection and classification of integrons and gene cassettes
Methods used to extract bacterial DNA and detect integrons were as previously described by our group.14,21 Briefly, integrons were detected using polymerase chain reaction (PCR), with primers targeting conserved regions of integron-encoded integrases intI1, intI2, and intI3.14,21 Integrase PCR products were subjected to restriction fragment length polymorphism (RFLP) analysis, using HinfI and RsaI to determine integron class as previously described.14 Gene cassette regions were amplified by PCR and characterised by sequencing and RFLP.14,21 Analysis of sequence data was performed using programs provided in WebANGIS, by the Australian National Genomic Information Service.
Top of page
Results
Resistance profiles
All strains tested were resistant to ampicillin, while 18 of 19 (95%) organisms were resistant to cefotaxime, 17 of 19 (89%) organisms were resistant to cephalexin, streptomycin, tobramycin and nitrofurantoin (Table 2). Most strains were susceptible to imipenem, with only 37 per cent of organisms resistant to this antibiotic (Table 2). All strains were resistant to at least 50 per cent of the range of antibiotics tested and one strain, Acinetobacter baumanii INSJ18, was resistant to all antibiotics tested (data not shown).
Table 2. Percentage of organisms resistant to antibiotics tested
Antibiotic |
Resistant organisms
% |
| Amikacin |
63 |
| Gentamicin |
79 |
| Kanamycin |
84 |
| Netilmicin |
68 |
| Streptomycin |
89 |
| Tobramycin |
89 |
| Ampicillin |
100 |
| Augmentin |
74 |
| Cefotaxime |
95 |
| Cefotetan |
47 |
| Cephalexin |
89 |
| Imipenem |
37 |
| Timentin |
84 |
| Nalidixic acid |
68 |
| Norfloxacin |
58 |
| Chloramphenicol |
84 |
| Nitrofurantoin |
89 |
| Sulphafurazole |
74 |
| Tetracycline |
79 |
| Trimethoprim |
74 |
Top of page
Integron detection and classification
Organisms were screened for the presence of integrase genes by PCR in order to determine the prevalence of integrons in the multi-resistant collection. Nine of 19 (47%) strains contained at least one integron. RFLP analysis revealed that six isolates contained a single class 1 integron, one strain contained two class 1 integrons, one strain contained both a class 1 and a class 2 integron, and in one other strain, a single class 2 integron was detected (Table 3). Class 3 integrons were not detected in this study.
Table 3. Integron cassette arrays and integron-associated antibiotic resistance profiles of integrase positive organisms
Strain |
Cassettes contained by integron 1† |
Cassettes contained by integron 2† |
Resistant to antibiotics* |
| AK |
CN |
K |
NET |
S |
TOB |
AMP |
SF |
W |
| P. mirabilis INSJ04 |
aacA4, aacC1, orfXa, orfXb, aadA1 |
dfrA1, sat1, aadA1‡ |
R |
R |
R |
R |
R |
R |
R |
R |
R |
| P. mirabilis INSJ10 |
dfrA1, sat1, aadA1‡ |
– |
R |
R |
R |
R |
R |
R |
R |
R |
R |
| Klebsiella sp. INSJ11 |
oxa1, aadA1 |
– |
S |
R |
R |
R |
R |
R |
R |
R |
R |
| K. pneumoniae INSJ16 |
aadB |
– |
R |
R |
R |
R |
S |
R |
R |
R |
S |
| P. mirabilis INSJ20 |
aacA4, oxa2, orfD |
aadB, aadA1 |
S |
R |
R |
R |
R |
R |
R |
R |
R |
| E. coli INSJ21 |
dfrA1, aadA1 |
– |
R |
S |
S |
S |
R |
S |
R |
R |
R |
| S. Typhimurium INS95 |
aadA2 |
– |
S |
R |
S |
S |
R |
R |
R |
R |
S |
| C. freundii INSTR2 |
NA |
– |
R |
S |
R |
R |
R |
R |
R |
R |
R |
| E. cloaceae INSTR5 |
aadA1 |
– |
R |
R |
R |
R |
R |
R |
R |
S |
S |
Top of page
Gene cassette characterisation
Analysis of the cassette regions revealed class 1 integrons contained between one and five different gene cassettes (Table 3). Class 2 integrons contained the three gene cassettes associated with Tn7, namely dfrA1 (trimethoprim resistance),22sat1 (streptothricin resistance)4 and aadA1 (streptomycin/spectinomycin resistance).23 A predominance (15/23) of gene cassettes were identified that confer resistance to the aminoglycosides (Table 3). These cassettes were identified as aadA1 and aadA2,24aadB (resistance to gentamicin, kanamycin and tobramycin),12aacA4 (resistance to amikacin, netilmicin and tobramycin),25 and aacC1 (resistance to gentamicin, astromicin and sisomicin).26 Gene cassettes conferring resistance to trimethoprim (dfrA1) and the ▀-lactams (oxa1 and oxa2) (i.e., oxacillin and ampicillin resistance)27,28 were also identified (Table 3). Resistance conferred by the gene cassette correlated with phenotypic resistance as determined by susceptibility testing (Table 3). In addition, all strains containing a class 1 integron with the exception of Enterobacter cloaceae INSTR5, were resistant to sulphonamides (Table 3). This sulfonamide resistance is probably due to the presence of a sul1 gene that is nearly always found downstream of the cassette array in class 1 integrons.
Top of page
Discussion
Integrons have been recognised as important contributors to antibiotic susceptibility profile of Gram-negative isolates.14,15,16,17,18,19 Nine of 19 (47%) multi-resistant Gram-negative clinical isolates contained at least one integron, and 2 of 19 (11%) strains contained two integrons. The proportion of strains in this collection of bacteria carrying integrons is comparable to other studies. For example, 49 per cent of 120 urinary isolates of Enterobacteriaceae in Sydney were found to carry integrons,14 52 per cent of 54 clinical isolates of E. coli in Taiwan,15 and 43 per cent of 163 Gram-negative isolates in European hospitals contained class 1 integrons.18
Integrons have not only been found in isolates from human infection. They have also been reported in environmental and animal isolates, for example, integrons have been reported in bacteria from diseased poultry,29 fish,30 pigs and cattle31 and retail ground meats.32 Thus, there is potential for the transfer of integron-carrying bacteria from these sources to humans.
Up to 50 per cent of multi-resistant strains of A. baumanii have been previously reported to carry class 1 and 2 integrons.33 However, integrons were not detected in the four strains of Acinetobacter baumanii in this study although they were resistant to most antibiotics tested (data not shown). These strains, along with the 53 per cent of bacteria that were integron negative, demonstrate that although integrons are significantly associated with a multi-resistance phenotype,14 multi-resistance in some isolates appears to be mediated by other mechanisms.
In 2000, an outbreak of shigellosis spread rapidly through a community of homosexual men in Sydney and was attributed to a Shigella sonnei biotype g strain.34 Retrospective PCR screening of outbreak strains by our group revealed that they all harboured class 2 integrons that contain resistance genes to streptomycin, streptothricin and trimethoprim.34 This study highlighted the need for improved control of the spread of resistance-carrying bacteria and demonstrates usefulness of molecular screening techniques for rapid identification of resistance genes. Information provided could have been used to alter the continuation of ineffective antibiotic treatments that occurred during the outbreak.
Integron-screening and gene cassette characterisation can potentially be utilised as a rapid PCR-based method of resistance profile analysis that allows the identification of genetic resistance determinants. Integrons are a marker for multi-resistance, hence integron screening can be used to predict phenotypic antibiotic resistance. The presence of integrons in clinical isolates is of concern due to their ability to capture further gene cassettes. This gives the host organism the potential to acquire resistance against a wide variety of antibiotics, since gene cassettes exist to nearly all classes of antibiotic. Additionally, integron-screening provides the potential for identification of new resistance gene cassettes, demonstrated by characterisation of two novel gene cassettes aadA5 and dfrA17 by our group in 2000.35 In the present study of Gram-negative multi-resistant bacteria, we have found that integrons contribute considerably to the resistance profiles of nearly 50 per cent of these organisms. This information complements antibiotic resistance surveillance programs, providing information on the molecular mechanisms of resistance in addition to elucidating means of resistance gene acquisition.
Top of page
Acknowledgements
We are grateful to Professor Sydney Bell, Dr Jeannette Pham and Dr Barrie Gatus, Antibiotic Reference Laboratory, Department of Microbiology (SEALS), Prince of Wales Hospital, for the provision of strains used in this study.
Top of page
References
1. Hall RM, Collis CM. Antibiotic resistance in Gram-negative bacteria-the role of gene cassettes and integrons. Drug Resist Updat 1998;1:109-119.
2. Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 1989;3:1669-1683.
3. Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol 1995;15:593-600.
4. Wohllenben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy PH, et al. On the evolution of Tn21-like multiresistant transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC-(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet 1989;217:202-208.
5. Bissonnette L, Champetier S, Buisson JP, Roy PH. Characterisation of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: Similarity of the product to transmembrane transport proteins. J Bacteriol 1991;173:4493-4502.
6. Hall RM, Brookes DE, Stokes HW. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol 1991;5:1941-1959.
7. Collis CM, Hall RM. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol 1992;6:2875-2885.
8. Collis CM, Hall RM. Site-specific deletion and rearrangement of integron insert genes catalysed by the integron DNA integrase. J Bacteriol 1992;174:1574-1585.
9. Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. Site-specific insertion of gene cassettes into integrons. Mol Microbiol 1993;9:41-52.
10. Recchia GD, Stokes HW, Hall RM. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res 1994;22:2071-2078.
11. Naas T, Mikami Y, Imai T, Poirel L, Nordmann P. Characterisation of In53, a class 1 plasmid-and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol 2001;183:235-249.
12. Cameron FH, Groot Obbink DJ, Ackerman VP, Hall RM. Nucleotide sequence of the AAD(2'') aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res 1986;14:8625-8635.
13. Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother 1995;39:155-162.
14. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother 2001;45:2658-2661.
15. Chang C, Chang L, Chang Y, Lee T, Chang S. Characterisation of drug gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J Med Microbiol 2000;49:1097-1102.
16. Lee JC, Oh JY, Cho JW, Park JC, Kim JM, Seol SY, et al. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J Antimicrob Chemother 2001;47:599-604.
Top of page
17. Levesque C, Piche L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 1995;39:185-191.
18. Martinez-Freijo P, Fluit AC, Schmitz FJ, Grek VSC, Verhoef J, Jones ME. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother 1998;42:689-696.
19. Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, et al. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J Clin Microbiol 1996;34:2909-2913.
20. Bell SM, Gatus BJ, Pham JN. Antibiotic susceptibility testing by the CDS method. A concise laboratory manual. Arthur Productions Pty Ltd, Sydney. NSW: The antibiotic reference laboratory, South Eastern Area Laboratory Services; 1999.
21. White PA, McIver CJ, Deng YM, Rawlinson WD. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol Lett 2000;182:265-269.
22. Fling ME, Richards C. The nucleotide sequence of the trimethoprim-resistant dihydrofolate reductase gene harbored by Tn7. Nucleic Acids Res 1983;11:5147-5158.
23. Sundstrom L, Radstrom P, Swedberg G, Skold O. Site-specific recombination promotes linkage between trimethoprim-and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet 1988;213:191-201.
24. Bito A, Susani M. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob. Agents Chemother 1994;38:1172-1175.
25. Tolmasky ME. Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 1990;24:218-226.
26. Tenover FC, Phillips KL, Gilbert T, Lockhart P, O'Hara PJ, Plorde JJ. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob Agents Chemother 1989;33:551-559.27. Ouellette M, Bissonnette L, Roy PH. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc Natl Acad Sci USA 1987;84:7378-7382.
28. Hall RM, Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res 1987;15:7491-7501.
29. Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, et al. Incidence and characterisation of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob Agents Chemother 1999;43:2925-2929.
30. Schmidt AS, Bruun MS, Larsen JL, Dalsgaard I. Characterisation of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicida isolates from various geographical areas. J Antimicrob Chemother 2001;47:735-743.
31. Sandvang D, Aarestrup FM. Characterisation of aminoglycoside resistance genes and class 1 integrons in procine and bovine gentamicin-resistant Escherichia coli. Microb Drug Resist 2000;6:19-27.
32. White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, et al. The isolation of antibiotic-resistant Salmonella from retail ground meats. N Engl J Med 2001;345:1147-1154.
33. Koeleman JG, Stoof J, Van der Bijl MW, Vadenbrouke-Grauls CMJE, Savelkoul PHM. Identification of epidemic strains of Acinetobacter baumanii by integrase gene PCR. J Clin Microbiol 2001;39:8-13.
34. McIver CJ, White PA, Jones LA, Karagiannis T, Harkness J, Marriott D, et al. Epidemic strains of Shigella sonnei Biotype g carrying integrons. J Clin Microbiol 2002;40:1538-1540.
35. White PA, Rawlinson WD. Current status of the aadA and dfr gene cassette families. FEMS Microbiol Lett 2001;182:265-269.
Top of page
Author affiliations
1. Virology Division, Department of Microbiology, SEALS, Prince of Wales Hospital, Randwick, New South Wales
2. Department of Microbiology and Immunology, School of Biotechnology and Biomolecular Sciences, Faculty of Science, The University of New South Wales, Randwick, New South Wales
3. School of Medical Sciences, Faculty of Medicine, The University of New South Wales, Randwick, New South Wales
Corresponding author: Dr Peter White, Virology Division, Department of Microbiology, SEALS, Prince of Wales Hospital, Randwick NSW 2031. Telephone: +61 2 9382 9096. Facsimile: +61 2 9398 4275. Email: whitepa@sesahs.nsw.gov.au
This article was published in Communicable Diseases Intelligence Volume 27 Suppl, May 2003.
